install.packages("duckdb")
library(duckdb)Just recently as part of this project I finally got around to putting some properly big data into an actual database and OMG! For the kind of situation I was in, I can’t recommend giving this a try enough.
Some background
The project in question as one component involves developing or at least exploring building :species distribution models for a large number of the bird species present in Aotearoa New Zealand. To that end we’ve obtained the latest eBird data collected for the New Zealand Bird Atlas. This is a phenomenal resource which includes the accumulated observations of thousands of citizen science volunteers, accumulated over several years.
The raw .txt file containing the observational data is a chunky 2.8GB with 7 million rows of data. Seven million rows isn’t so bad, right? Right, it really isn’t that bad. There is even an R package (of course there is), cheekily called auk1, for massaging the raw data down to the data you actually want.
In my case relevant data pertain only to the NZ Bird Atlas effort, and to complete checklists as only these provide the absence data required for occupancy modelling. The complexity of the data makes running auk to filter the raw data down slow, but it’s a one-time-only operation, so that’s OK, and now I have a 3.7 million row table, and we’re in business, no need to worry about setting anything up.
The problem comes when I have to blow those 3.7 million rows back up again for occupancy modelling to 110 million rows.
Wait, what? 110 million rows?!
Yes, 110 million rows.
How it works is that each species (a little over 300) requires an entry in each complete checklist (around 360,000 of these) recording whether that species was observed (present) or not (absent) in that checklist. That results in 110 million row table. You can keep the two tables separate but then you have to keep joining them every time you go to use them. And if you save the 110 million table to disk it takes up around 8GB, and also takes a noticeable length of time to open and close for analysis in R.
It was about this time that I thought I should consider my options.
Enter DuckDB
DuckDB is an easy to install columnar database that you can drive using :SQL. It’s particularly easy to install because the R and Python APIs come bundled with the database itself. So, if you don’t want to, you don’t even have to install it, your platform of choice will do that for you.
Anyway, if you do install it, which I did, so I could poke it around a little before going further, then to start it up from the command line type
% duckdbAnd if you want to create a new database in the folder you are running from then it’s
% duckdb my-new-database.dbThe file extension is optional. Once in the session you can stash an existing CSV file in the database as a table with the command (D is the DuckDB command line prompt):
D CREATE TABLE letters AS FROM 'letters.csv';and to see the results of your handiwork:
D SELECT * FROM letters;
┌─────────┬───────┬─────────┐
│ column0 │ id │ letter │
│ int64 │ int64 │ varchar │
├─────────┼───────┼─────────┤
│ 1 │ 1 │ a │
│ 2 │ 2 │ b │
│ 3 │ 3 │ c │
│ 4 │ 4 │ d │
│ 5 │ 5 │ e │
│ 6 │ 6 │ f │
│ 7 │ 7 │ g │
│ 8 │ 8 │ h │
│ 9 │ 9 │ i │
│ 10 │ 10 │ j │
├─────────┴───────┴─────────┤
│ 10 rows 3 columns │
└───────────────────────────┘
D Satisfied it was this easy, I typed .exit to shut DuckDB down and moved on to consider how to use DuckDB from R. I should mention at this point that I’ve bounced off PostgreSQL a couple of times in the past when considering using it in classroom situations because it’s just not as easy to get into as this.
DuckDB in R
The R package you need is duckdb, so
and you are ready to go (no other installation of DuckDB required).
Now if you have a giant dataframe called say my_giant_df, that you need to deal with, open a connection to a new database (or an existing one if you’ve been here before) with
con <- dbConnect(duckdb(), "my-giant-dataframe.db")and write your dataframe into it as a table called giant_df with
dbWriteTable(con, "giant_df", my_giant_df)If that’s all you plan on doing then you should shut down the connection
dbDisconnect(con, shutdown = TRUE)When I did this I was agreeably surprised to find that my 8GB file had shrunk down to a mere 750MB.
But there’s more. The reason I went down this route at all is that I generally only want to work with the data for one bird species at a time — data which come in handy packets of only 360,000 rows or so. Here’s how that works in practice. First open a connection to the database
con <- dbConnect(duckdb(), dbdir = str_glue("the-birds.db"))The database has a table called observations containing the aforementioned 110 million rows. Each row includes among other things the common_name of a bird. We can get a vector containing those using
common_names <- dbGetQuery(con, "SELECT DISTINCT common_name FROM observations") |>
pull(common_name) |>
sort()Now we can iterate over each species by doing
for (common_name in common_names) {
sql_name <- str_replace_all(common_name, "'", "''")
query <- str_glue(str_glue("SELECT * FROM observations WHERE common_name = '{sql_name}'"))
this_bird_df <- dbGetQuery(con, query)
# ...
# do stuff with this_bird_df
# ...
}The only wrinkles here, for those paying attention, are using str_glue from the stringr package to form the SQL query I need, and related to that a str_replace_all to double up any single-quotes ' that happen to appear in those common names to '' so that they can be passed into an SQL query.
In general you query the database using dbGetQuery(<connection>, <SQL>) and you can execute a command with dbExecute(con, <command>).
Of course, you have to know a bit of SQL, but for this kind of simple (local) data warehousing, there’s nothing you are likely to need that a quick google DuckDuckGo search won’t unearth.
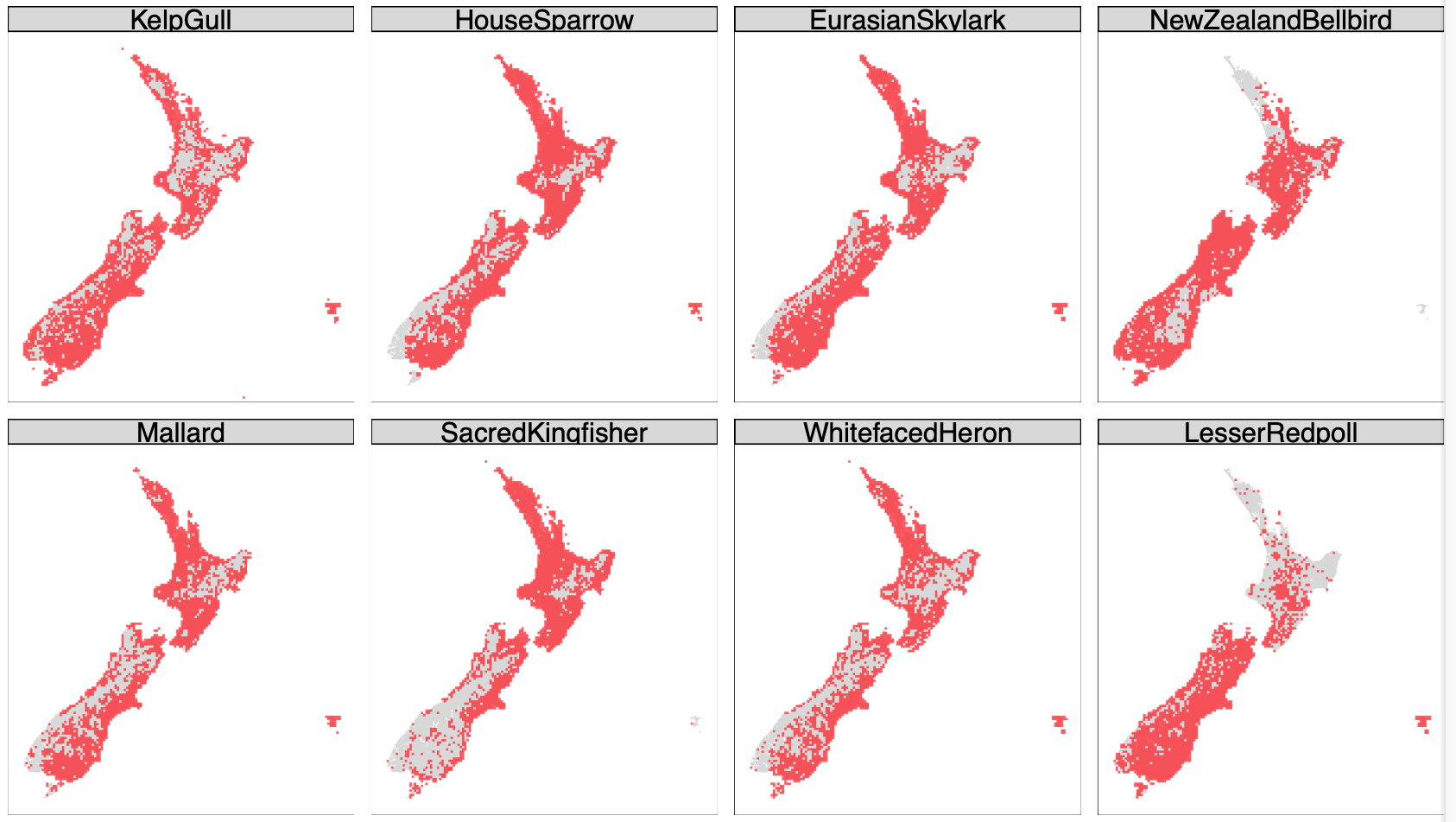
This approach enabled me to iterate over all 300 species in the data and assemble a ‘mini-atlas’ of ggplot maps of each bird’s range in under a minute (snippet below), which is about how long it was previously taking R just to open the 8GB CSV file. Not to mention that the giant data table is never in working memory, only the chunks I need one at a time.
It’s safe to say, I’ll be using DuckDB a lot in many projects to come. There seem to be some wrinkles in relation to handling spatial data, specifically from R’s sf package but there’s a package for that,2 and it’s nothing to get too alarmed about.
Footnotes
A very nerdy deep cut from the Cornell Ornithology Lab. Respect.↩︎
‘We have a package for that’ should be R’s tagline.↩︎